Multilayer graphene pores with 0 and 60 degrees relative rotation of flakes


href=”https://petr-kral.com/wp-content/uploads/2023/12/multi1.avi”
href=”https://petr-kral.com/wp-content/uploads/2023/12/multi2.avi”
Ion passage in multilayer graphene pores with 0 and 60 degrees rotation
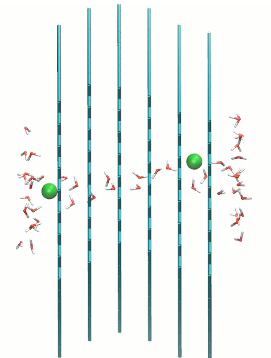
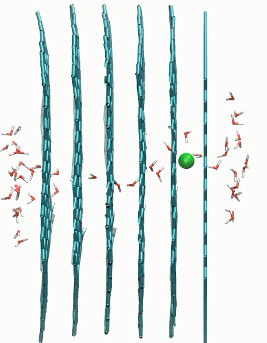
href=”https://petr-kral.com/wp-content/uploads/2023/12/multi3.avi”
href=”https://petr-kral.com/wp-content/uploads/2023/12/multi4.avi”
L and D-leucine passing multilayer graphene pores with 60 degrees rotation
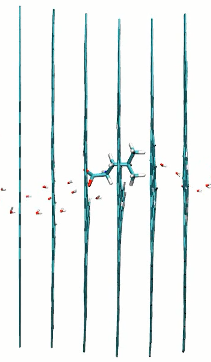
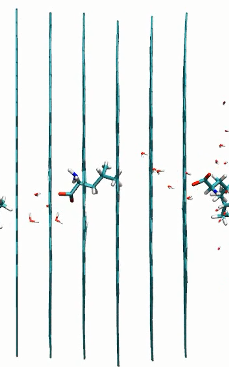
href=”https://petr-kral.com/wp-content/uploads/2023/12/multi5-L-leucine.avi
href=”https://petr-kral.com/wp-content/uploads/2023/12/multi6-D-leucine.avi
Self-assembly of nanoparticles within a wanochannel
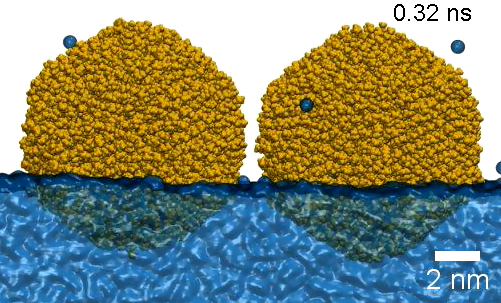
https://petr-kral.com/wp-content/uploads/2023/12/filaments.avi
Transient byproducts clustering on wet-etched Silicon nanopillars

https://petr-kral.com/wp-content/uploads/2023/12/etching.avi
PEG(350)ylated organomimetic cluster nanomolecules (OCNs) in water
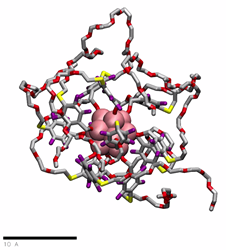
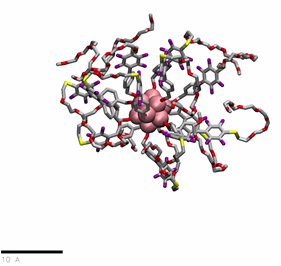
href=”https://petr-kral.com/wp-content/uploads/2023/12/G1-PEG350.mp4
href=”https://petr-kral.com/wp-content/uploads/2023/12/G2-PEG350.mp4
PEG(750)ylated OCNs in water
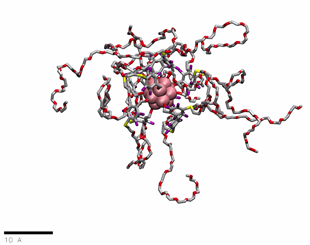
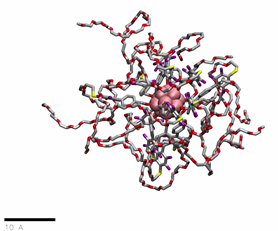
href=”https://petr-kral.com/wp-content/uploads/2023/12/G1-PEG750.mp4
href=”https://petr-kral.com/wp-content/uploads/2023/12/G2-PEG750.mp4
PEG(2000)ylated OCNs in water
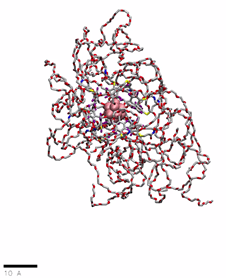
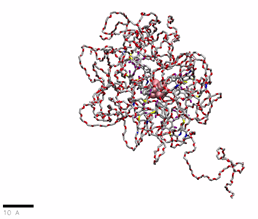
href=”https://petr-kral.com/wp-content/uploads/2023/12/G1-PEG2000.mp4
href=”https://petr-kral.com/wp-content/uploads/2023/12/G2-PEG2000.mp4
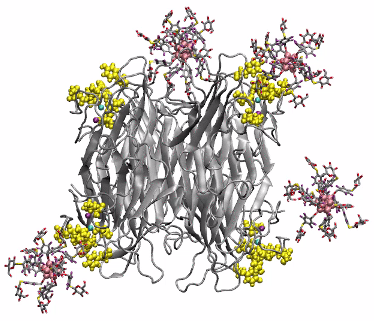
Multivalent binding of glycosylated OCNs to lectin Concanavalin A
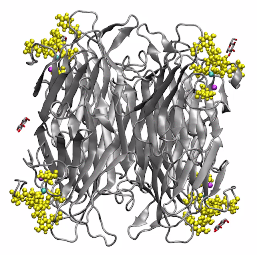
href=”https://petr-kral.com/wp-content/uploads/2023/12/sugar.mp4
href=”https://petr-kral.com/wp-content/uploads/2023/12/sugar-NP.mp4
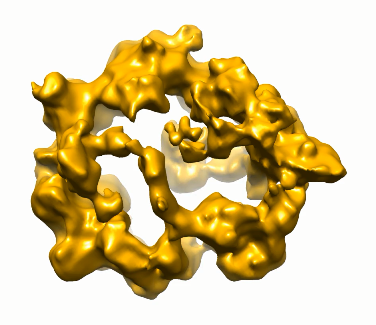
3D structure of nanoshells from CdS NPs obtained by TEM tomography
https://petr-kral.com/wp-content/uploads/2023/12/capsule1.mp4
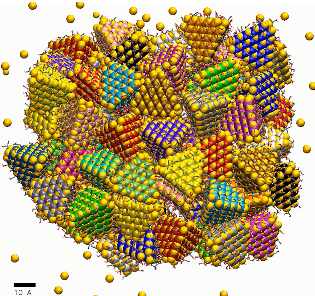
MD simulations of a nanoshell from ligated CdS NPs with charge q=2e
https://petr-kral.com/wp-content/uploads/2023/12/capsule2.mp4
MD simulations of a nanoshell from naked CdS NPs with charge q=0.6e

https://petr-kral.com/wp-content/uploads/2023/12/capsule3.mp4
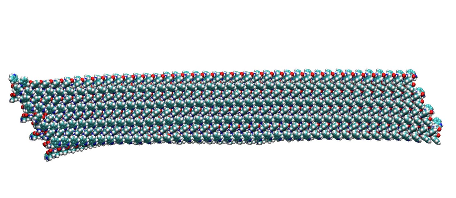
MD simulation of tryptophan bilayer ribbon – longitudinal
https://petr-kral.com/wp-content/uploads/2023/12/tryptophan-L.avi
MD simulation of tryptophan bilayer ribbon – transversal
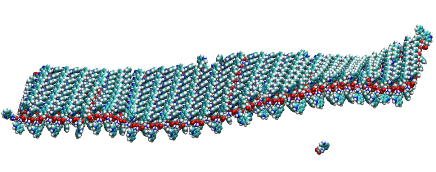
https://petr-kral.com/wp-content/uploads/2023/12/tryptophan-T.avi
In situ TEM movie showing formation of NP chain (100 f/s) – low linker concentration
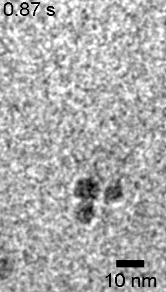
https://petr-kral.com/wp-content/uploads/2023/12/lowTEM.avi
MD simulation showing formation of NP chain (4 ns) – low linker concentration
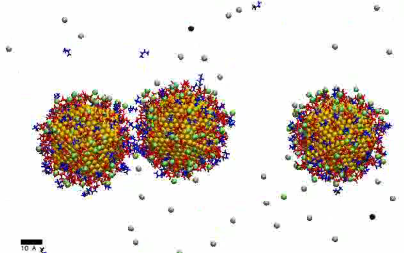
https://petr-kral.com/wp-content/uploads/2023/12/lowMD.avi
In situ TEM movie showing formation of branched NP network (25 f/s) – high linker concentration
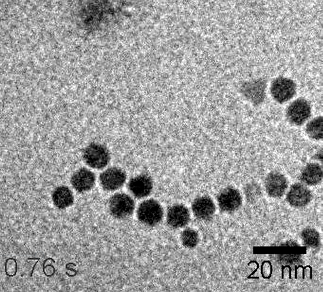
https://petr-kral.com/wp-content/uploads/2023/12/lowTEM.avi
MD simulation showing formation of NP chain (10 ns) – high linker concentration
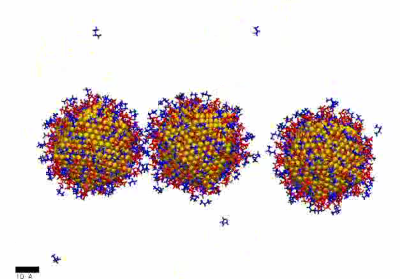
https://petr-kral.com/wp-content/uploads/2023/12/highMD-lin.avi
MD simulation showing formation of branched NP network (35 ns) – high linker concentration
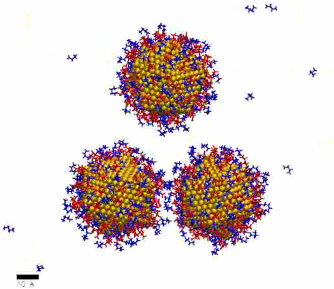
https://petr-kral.com/wp-content/uploads/2023/12/highMD-bra.avi
cis-azobenzene-ligated nanoparticles in toluene solvent and water solute
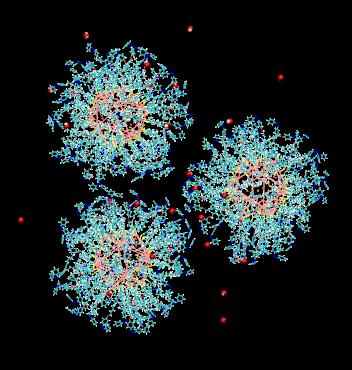
https://petr-kral.com/wp-content/uploads/2023/12/3cis.gif
trans-azobenzene-ligated nanoparticles in toluene solvent and water solute
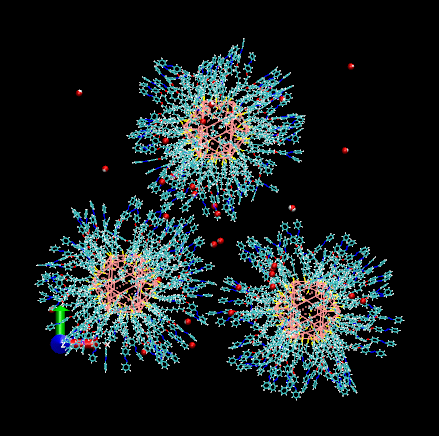
https://petr-kral.com/wp-content/uploads/2023/12/3trans.gif
Solvation of 20 water cluser between 4 cis-azobenzene-ligated nanoparticles
https://petr-kral.com/wp-content/uploads/2023/12/20waters.gif
Release of water in vacuum during a cis-trans ligand-type switching
https://petr-kral.com/wp-content/uploads/2023/12/cis-trans-vac.gif
Formation of helices of superparamagnetic nanocubes
https://petr-kral.com/wp-content/uploads/2023/12/magnet1.gif
Self-assembly of superstructures from a randomly positioned nanocubes
https://petr-kral.com/wp-content/uploads/2023/12/magnet2.gif
Coupling of F anion to coronene (center)
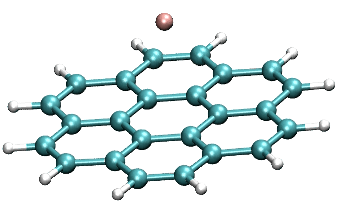
https://petr-kral.com/wp-content/uploads/2023/12/coro-F-center.avi
Coupling of F anion to coronene (side)
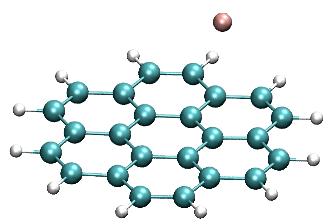
https://petr-kral.com/wp-content/uploads/2023/12/coro-F-side.avi
Coupling of Na cation to coronene (center)
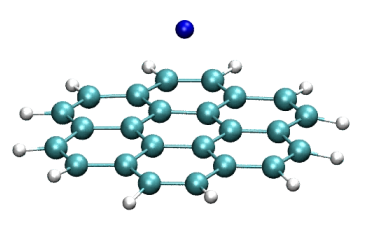
https://petr-kral.com/wp-content/uploads/2023/12/coro-Na-center.avi
Coupling of F anion to a triangular flake (center)
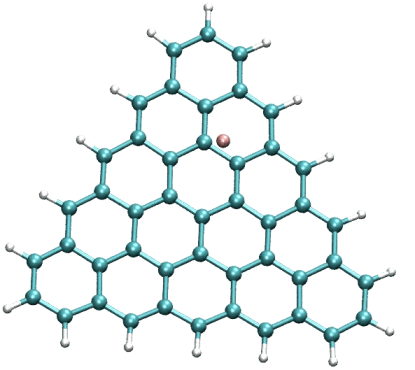
https://petr-kral.com/wp-content/uploads/2023/12/F-zig-cen-bond.avi
Coupling of F anion to a triangular flake (edge)
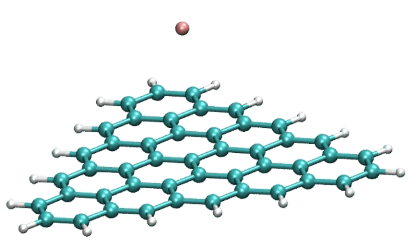
https://petr-kral.com/wp-content/uploads/2023/12/zigzag-F-edge.avi
Dragging of small water droplets on vibrated nanotubes

https://petr-kral.com/wp-content/uploads/2023/12/dropS.avi
Dragging of large water droplets on vibrated nanotubes
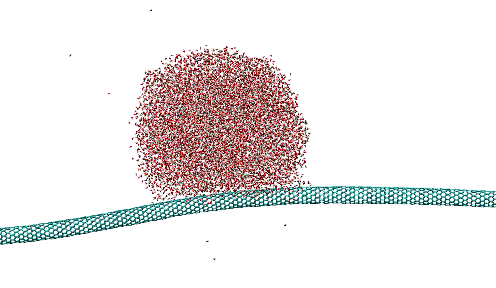
https://petr-kral.com/wp-content/uploads/2023/12/dropL.mpg
Nesting of molecules on functionalized graphene
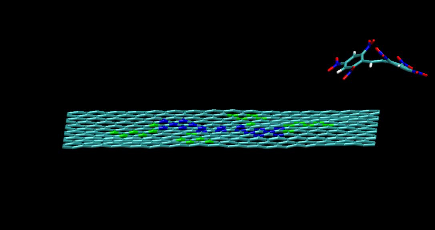
https://petr-kral.com/wp-content/uploads/2023/12/nest.mpg
Controlled self-assembly of double-headed lipid micelles

https://petr-kral.com/wp-content/uploads/2023/12/double-headed.avi
Controlled self-assembly of single-headed lipid micelles

https://petr-kral.com/wp-content/uploads/2023/12/single-headed.avi
Self-assembly of hydrated graphene-membrane superstructures
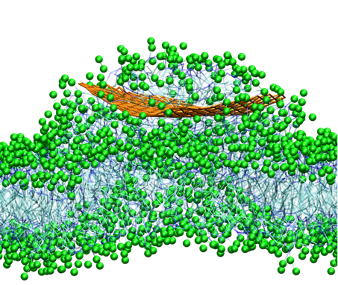
https://petr-kral.com/wp-content/uploads/2023/12/grabim.mpg
Rolling of graphene nanoribbon on carbon nanotube
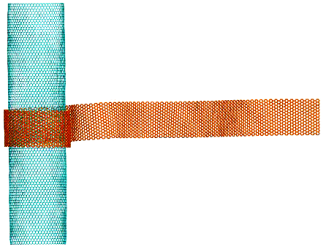
https://petr-kral.com/wp-content/uploads/2023/12/roll.mpg
Helical rolling of graphene nanoribbon on carbon nanotube
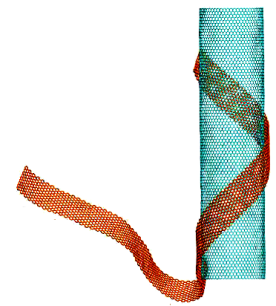
https://petr-kral.com/wp-content/uploads/2023/12/helix-out.mpg
Helical rolling of graphene nanoribbon inside carbon nanotube
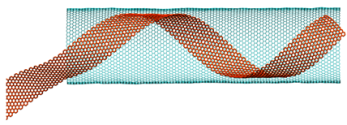
https://petr-kral.com/wp-content/uploads/2023/12/helix-in.mpg
Helical rolling of two graphene nanoribbons inside carbon nanotube

https://petr-kral.com/wp-content/uploads/2023/12/double-helix.mpg
Knot formation of graphene nanoribbon on carbon nanotube

https://petr-kral.com/wp-content/uploads/2023/12/knot.mpg
Self-assembly of graphene flake
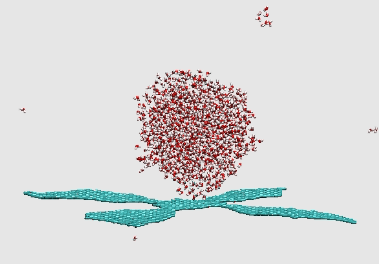
https://petr-kral.com/wp-content/uploads/2023/12/flower.avi
Sliding self-assembly of graphene stripe

https://petr-kral.com/wp-content/uploads/2023/12/sliding.avi
Rolling self-assembly of graphene stripe
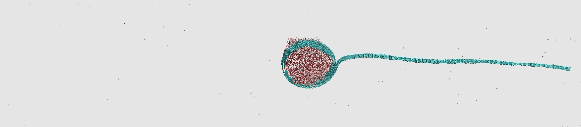
https://petr-kral.com/wp-content/uploads/2023/12/rolling.avi
Rolling of nanorods on watter by light
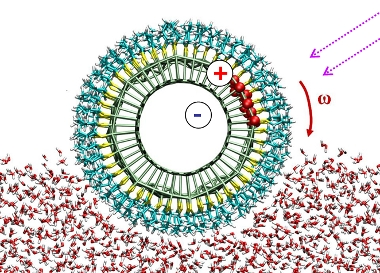
https://petr-kral.com/wp-content/uploads/2023/12/roller.avi
Twisting of hydrated nanotube-peptide junction
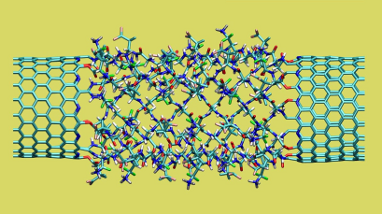
https://petr-kral.com/wp-content/uploads/2023/12/barcone.avi
Passage of Na+ ion through F-N-pore in graphene
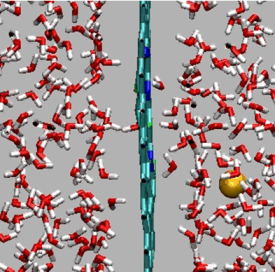
https://petr-kral.com/wp-content/uploads/2023/12/Na.avi
Passage of Cl- ion through H-pore in graphene
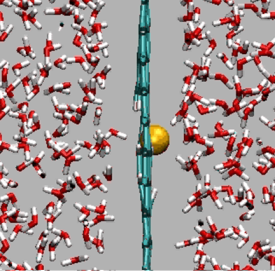
https://petr-kral.com/wp-content/uploads/2023/12/Cl.avi
Rotary tunneling motor with 3 blades
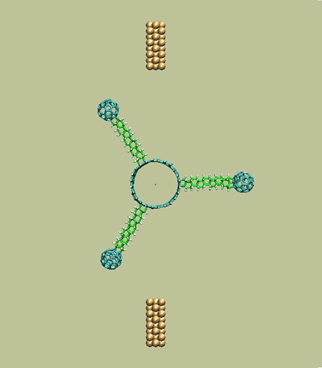
https://petr-kral.com/wp-content/uploads/2023/12/f3.avi
Rotary tunneling motor with 6 blades
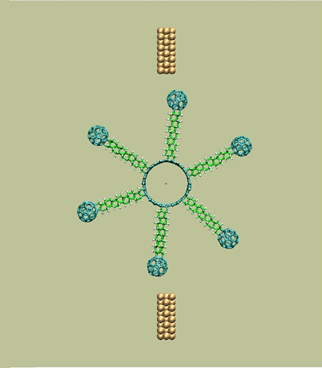
https://petr-kral.com/wp-content/uploads/2023/12/f6.avi
Rotary tunneling motor with dual-blades
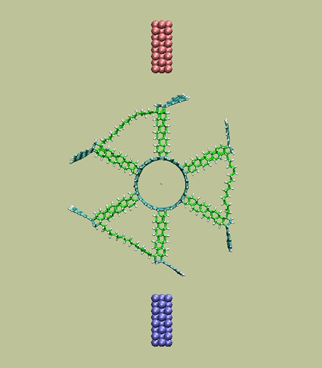
https://petr-kral.com/wp-content/uploads/2023/12/dip.avi
Dragging of 400 waters by Na ion on CNT
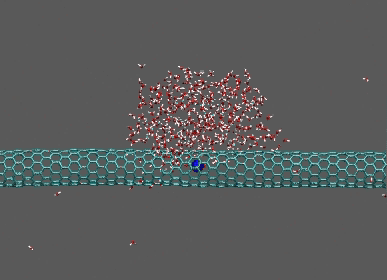
https://petr-kral.com/wp-content/uploads/2023/12/drop.avi
Dragging of 10000 waters by two Na ions on two CNTs
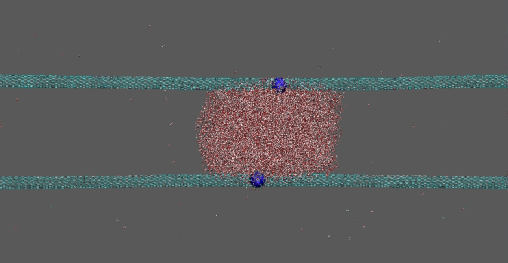
https://petr-kral.com/wp-content/uploads/2023/12/bigd.avi
Dragging of 400 waters by Na ion on BNT
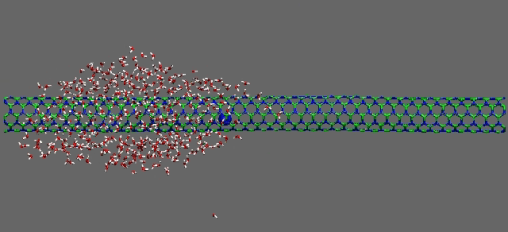
https://petr-kral.com/wp-content/uploads/2023/12/BNT5-5-400.avi
Molecular drag by separately flowing water
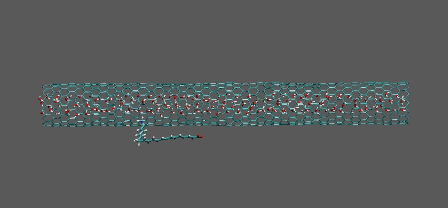
https://petr-kral.com/wp-content/uploads/2023/12/push.avi
Hydrophobic surface propeller

https://petr-kral.com/wp-content/uploads/2023/12/surf-pho.avi
Hydrophillic surface propeller

https://petr-kral.com/wp-content/uploads/2023/12/surf-phi.avi
Hydrophobic bulk propeller

https://petr-kral.com/wp-content/uploads/2023/12/bulk-pho.avi
Piece of GFP docked on the ligand-modified graphene
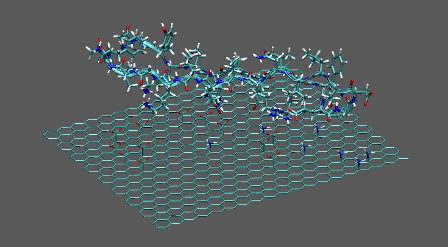
https://petr-kral.com/wp-content/uploads/2023/12/dop-pept.avi
Active catalyst on the right-twisted nanotube

https://petr-kral.com/wp-content/uploads/2023/12/catal-act.avi
Passive catalyst on the left-twisted nanotube
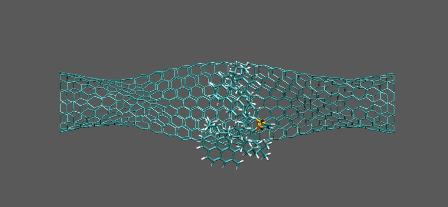
https://petr-kral.com/wp-content/uploads/2023/12/catal-pass.avi
|